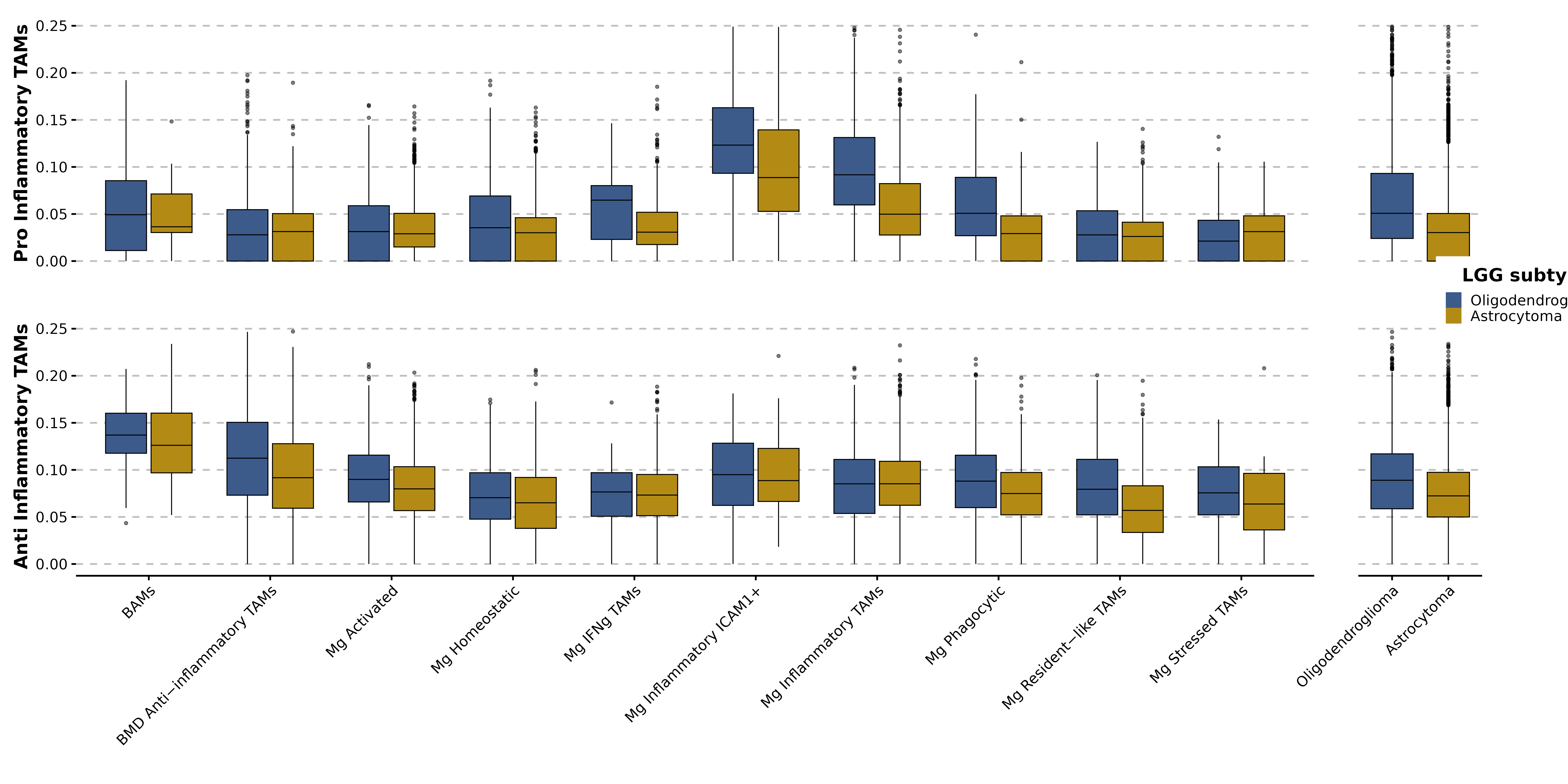
p1 <- SCpubr::do_BoxPlot(sample,
feature = "Pro_Inflammatory",
group.by = "final_annotation",
split.by = "subtype",
colors.use = scale.subtype.short,
font.size = 20,
axis.text.face = "plain",
legend.position = "right",
legend.title = "LGG subtype") +
ggplot2::ylab("Pro Inflammatory TAMs") +
ggplot2::xlab("") +
ggplot2::scale_y_continuous(limits = c(0, 0.25)) +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.line.x = ggplot2::element_blank(),
legend.position = "none")
p2 <- SCpubr::do_BoxPlot(sample,
feature = "Anti_Inflammatory",
group.by = "final_annotation",
split.by = "subtype",
colors.use = scale.subtype.short,
font.size = 20,
axis.text.face = "plain",
legend.position = "right",
legend.title = "LGG subtype") +
ggplot2::scale_y_continuous(limits = c(0, 0.25)) +
ggplot2::ylab("Anti Inflammatory TAMs") +
ggplot2::xlab("")
p3 <- SCpubr::do_BoxPlot(sample,
feature = "Pro_Inflammatory",
group.by = "subtype",
colors.use = scale.subtype.short,
font.size = 20,
axis.text.face = "plain",
legend.position = "right",
legend.title = "LGG subtype") +
ggplot2::scale_y_continuous(limits = c(0, 0.25)) +
ggplot2::ylab("") +
ggplot2::xlab("") +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.line.x = ggplot2::element_blank(),
axis.text.y = ggplot2::element_blank(),
axis.ticks.y = ggplot2::element_blank(),
axis.line.y = ggplot2::element_blank(),
legend.position = "none")
p4 <- SCpubr::do_BoxPlot(sample,
feature = "Anti_Inflammatory",
group.by = "subtype",
colors.use = scale.subtype.short,
font.size = 20,
axis.text.face = "plain",
legend.position = "right",
legend.title = "LGG subtype") +
ggplot2::scale_y_continuous(limits = c(0, 0.25)) +
ggplot2::ylab("") +
ggplot2::xlab("") +
ggplot2::theme(axis.text.y = ggplot2::element_blank(),
axis.ticks.y = ggplot2::element_blank(),
axis.line.y = ggplot2::element_blank())
layout <- "AAAAAAAAAAAAAAAAAAAACCE
BBBBBBBBBBBBBBBBBBBBDDE"
p <- patchwork::wrap_plots(A = p1,
B = p2,
C = p3,
D = p4,
E = patchwork::guide_area(),
guides = "collect",
design = layout)