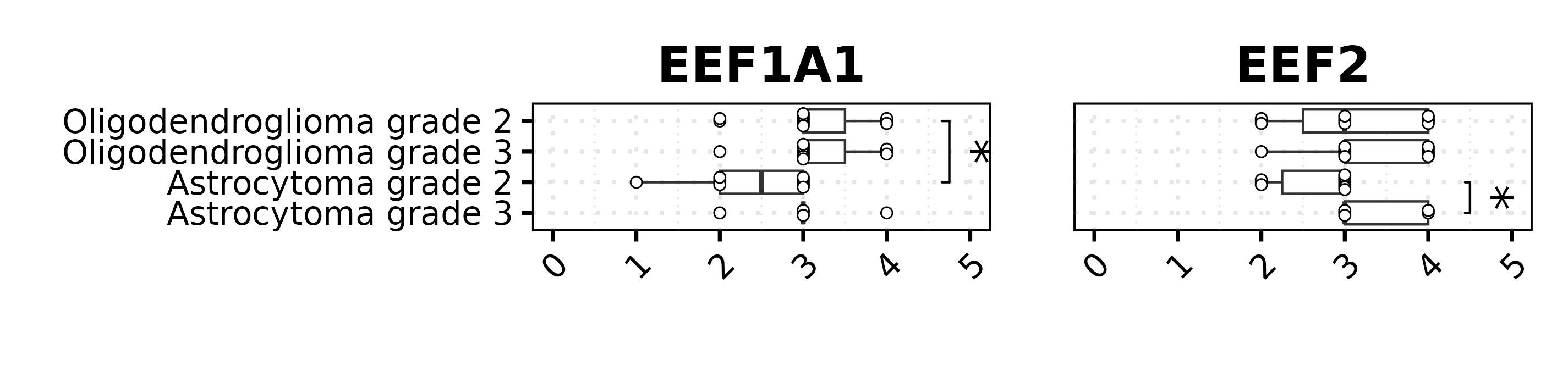
p1 <- data[data$Marker == "EEF1A1", ] %>%
ggplot2::ggplot(mapping = ggplot2::aes(y = forcats::fct_rev(.data$Type),
x = .data$Score)) +
ggplot2::geom_boxplot(na.rm = TRUE) +
ggbeeswarm::geom_beeswarm(size = 2,
color = "black",
fill = "white",
shape = 21,
stroke = 0.5,
cex = 2,
na.rm = TRUE) +
ggsignif::geom_signif(test = "wilcox.test",
comparisons = list(c("Oligodendroglioma grade 2", "Astrocytoma grade 2")),
map_signif_level = TRUE,
margin_top = 0.25,
na.rm = TRUE,
size = 0.5,
textsize = 8) +
ggplot2::labs(y = "",
x = "",
title = "EEF1A1") +
ggplot2::theme_minimal(base_size = 18) +
ggplot2::scale_x_continuous(limits = c(0, 5)) +
ggplot2::theme(plot.title = ggplot2::element_text(face = "bold",
hjust = 0.5,
vjust = 0),
plot.subtitle = ggplot2::element_text(face = "bold", hjust = 0),
plot.caption = ggplot2::element_text(face = "plain", hjust = 1),
legend.text = ggplot2::element_text(face = "plain"),
legend.title = ggplot2::element_text(face = "bold"),
plot.title.position = "panel",
plot.caption.position = "plot",
text = ggplot2::element_text(family = "sans"),
legend.justification = "center",
legend.position = "bottom",
axis.title.x = ggplot2::element_text(color = "black", face = "bold", hjust = 0.5),
axis.title.y.left = ggplot2::element_text(color = "black", face = "bold", angle = 90),
axis.title.y.right = ggplot2::element_blank(),
axis.text.y.left = ggplot2::element_text(color = "black",
face = "plain"),
axis.text.y.right = ggplot2::element_blank(),
axis.ticks.x.bottom = ggplot2::element_line(color = "black"),
axis.ticks.y.right = ggplot2::element_blank(),
axis.ticks.y.left = ggplot2::element_line(color = "black"),
axis.text.x.bottom = ggplot2::element_text(color = "black",
face = "plain",
angle = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["angle"]],
hjust = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["hjust"]],
vjust = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["vjust"]]),
strip.text.x = ggplot2::element_text(face = "bold",
color = "black"),
strip.text.y = ggplot2::element_blank(),
panel.border = ggplot2::element_rect(color = "black", fill = NA),
panel.grid = ggplot2::element_line(color = "grey90", linetype = "dotted"),
plot.margin = ggplot2::margin(t = 10, r = 10, b = 10, l = 0),
plot.background = ggplot2::element_rect(fill = "white", color = "white"),
panel.background = ggplot2::element_rect(fill = "white", color = "black", linetype = "solid"),
legend.background = ggplot2::element_rect(fill = "white", color = "white"))
p2 <- data[data$Marker == "EEF2", ] %>%
ggplot2::ggplot(mapping = ggplot2::aes(y = forcats::fct_rev(.data$Type),
x = .data$Score)) +
ggplot2::geom_boxplot(na.rm = TRUE) +
ggbeeswarm::geom_beeswarm(size = 2,
color = "black",
fill = "white",
shape = 21,
stroke = 0.5,
cex = 2,
na.rm = TRUE) +
ggsignif::geom_signif(test = "wilcox.test",
comparisons = list(c("Astrocytoma grade 2", "Astrocytoma grade 3")),
map_signif_level = TRUE,
margin_top = 0.25,
na.rm = TRUE,
size = 0.5,
textsize = 8) +
ggplot2::labs(y = "", x = "", title = "EEF2") +
ggplot2::theme_minimal(base_size = 18) +
ggplot2::scale_x_continuous(limits = c(0, 5)) +
ggplot2::theme(plot.title = ggplot2::element_text(face = "bold",
hjust = 0.5,
vjust = 0),
plot.subtitle = ggplot2::element_text(face = "bold", hjust = 0),
plot.caption = ggplot2::element_text(face = "plain", hjust = 1),
legend.text = ggplot2::element_text(face = "plain"),
legend.title = ggplot2::element_text(face = "bold"),
plot.title.position = "panel",
plot.caption.position = "plot",
text = ggplot2::element_text(family = "sans"),
legend.justification = "center",
legend.position = "bottom",
axis.title.x = ggplot2::element_text(color = "black", face = "bold", hjust = 0.5),
axis.title.y.left = ggplot2::element_text(color = "black", face = "bold", angle = 90),
axis.title.y.right = ggplot2::element_blank(),
axis.text.y.left = ggplot2::element_blank(),
axis.text.y.right = ggplot2::element_blank(),
axis.ticks.x.bottom = ggplot2::element_line(color = "black"),
axis.ticks.y.right = ggplot2::element_blank(),
axis.ticks.y.left = ggplot2::element_blank(),
axis.text.x.bottom = ggplot2::element_text(color = "black",
face = "plain",
angle = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["angle"]],
hjust = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["hjust"]],
vjust = SCpubr:::get_axis_parameters(angle = 45, flip = TRUE)[["vjust"]]),
strip.text.x = ggplot2::element_text(face = "bold",
color = "black"),
strip.text.y = ggplot2::element_blank(),
panel.border = ggplot2::element_rect(color = "black", fill = NA),
panel.grid = ggplot2::element_line(color = "grey90", linetype = "dotted"),
plot.margin = ggplot2::margin(t = 10, r = 10, b = 10, l = 0),
plot.background = ggplot2::element_rect(fill = "white", color = "white"),
panel.background = ggplot2::element_rect(fill = "white", color = "black", linetype = "solid"),
legend.background = ggplot2::element_rect(fill = "white", color = "white"))
p <- patchwork::wrap_plots(A = p1,
B = p2,
design = "AB")