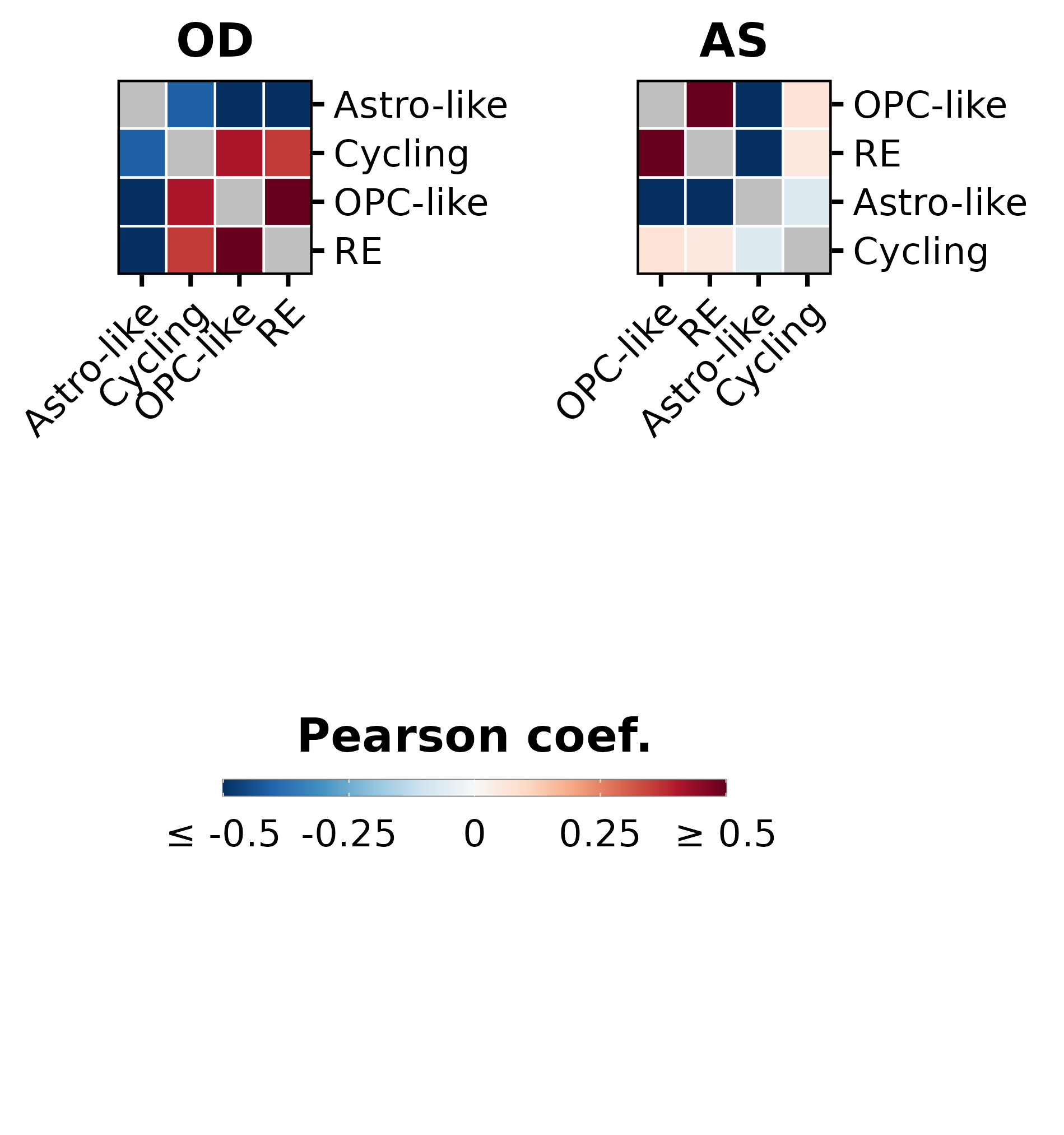
p1 <- SCpubr::do_CorrelationPlot(sample.oligo,
mode = "hvg",
assay = "ATAC",
max.cutoff = 0.5,
min.cutoff = -0.5,
font.size = 20,
legend.length = 15,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2,
axis.text.face = "plain",
legend.position = "none") +
ggplot2::xlab("OD")
p2 <- SCpubr::do_CorrelationPlot(sample.astro,
mode = "hvg",
assay = "ATAC",
max.cutoff = 0.5,
min.cutoff = -0.5,
font.size = 20,
legend.length = 15,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2,
axis.text.face = "plain") +
ggplot2::xlab("AS")
p <- patchwork::wrap_plots(A = p1, B = p2, C = patchwork::guide_area(),design = "AB
CC", guides = "collect")