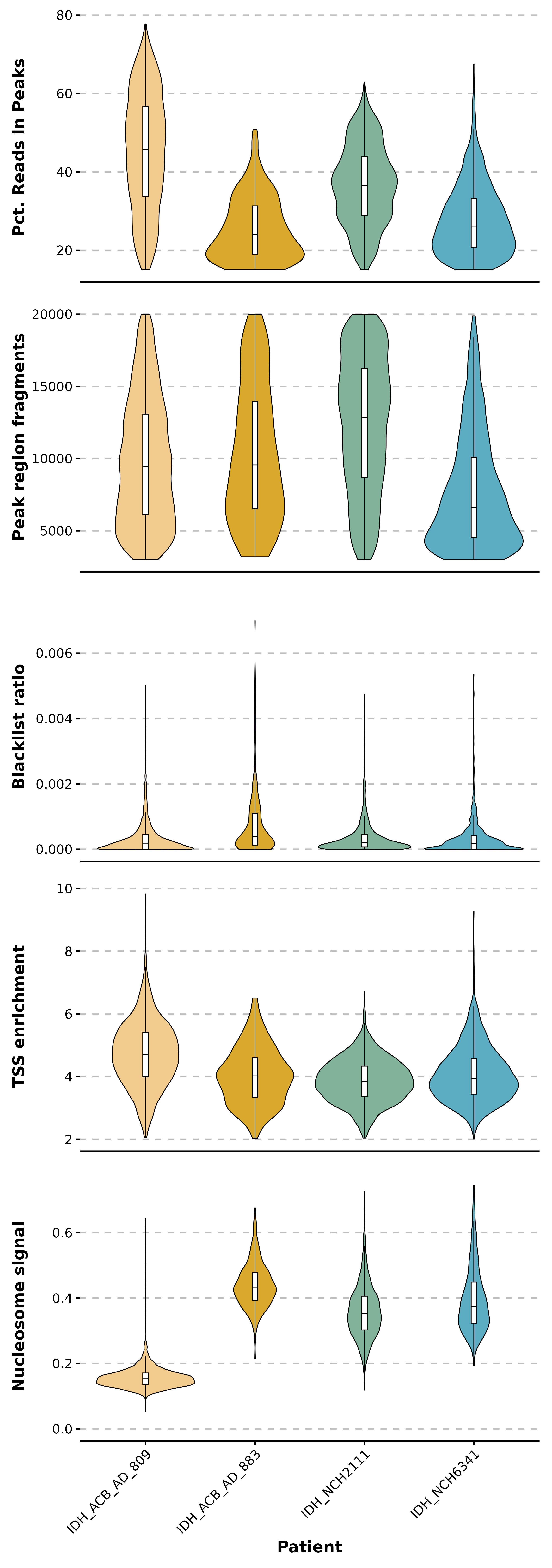
p1 <- SCpubr::do_ViolinPlot(sample = sample,
group.by = "orig.ident",
features = "percentage_reads_in_peaks",
colors.use = scale.ident,
boxplot_width = 0.05,
font.size = 20,
axis.text.face = "plain") +
ggplot2::labs(y = "Pct. Reads in Peaks",
x = "") +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.title.x = ggplot2::element_blank())
p2 <- SCpubr::do_ViolinPlot(sample = sample,
group.by = "orig.ident",
features = "peak_region_fragments",
colors.use = scale.ident,
boxplot_width = 0.05,
font.size = 20,
axis.text.face = "plain") +
ggplot2::labs(y = "Peak region fragments",
x = "") +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.title.x = ggplot2::element_blank())
p3 <- SCpubr::do_ViolinPlot(sample = sample,
group.by = "orig.ident",
features = "blacklist_ratio",
colors.use = scale.ident,
boxplot_width = 0.05,
font.size = 20,
axis.text.face = "plain") +
ggplot2::labs(y = "Blacklist ratio",
x = "") +
ggplot2::scale_y_continuous(limits = c(0, 0.0075)) +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.title.x = ggplot2::element_blank())
p4 <- SCpubr::do_ViolinPlot(sample = sample,
group.by = "orig.ident",
features = "TSS.enrichment",
colors.use = scale.ident,
boxplot_width = 0.05,
font.size = 20,
axis.text.face = "plain") +
ggplot2::labs(y = "TSS enrichment",
x = "") +
ggplot2::theme(axis.text.x = ggplot2::element_blank(),
axis.ticks.x = ggplot2::element_blank(),
axis.title.x = ggplot2::element_blank())
p5 <- SCpubr::do_ViolinPlot(sample = sample,
group.by = "orig.ident",
features = "nucleosome_signal",
colors.use = scale.ident,
boxplot_width = 0.05,
font.size = 20,
axis.text.face = "plain") +
ggplot2::labs(y = "Nucleosome signal",
x = "Patient") +
ggplot2::scale_y_continuous(limits = c(0, 0.75))
p <- p1 / p2 / p3 / p4 / p5