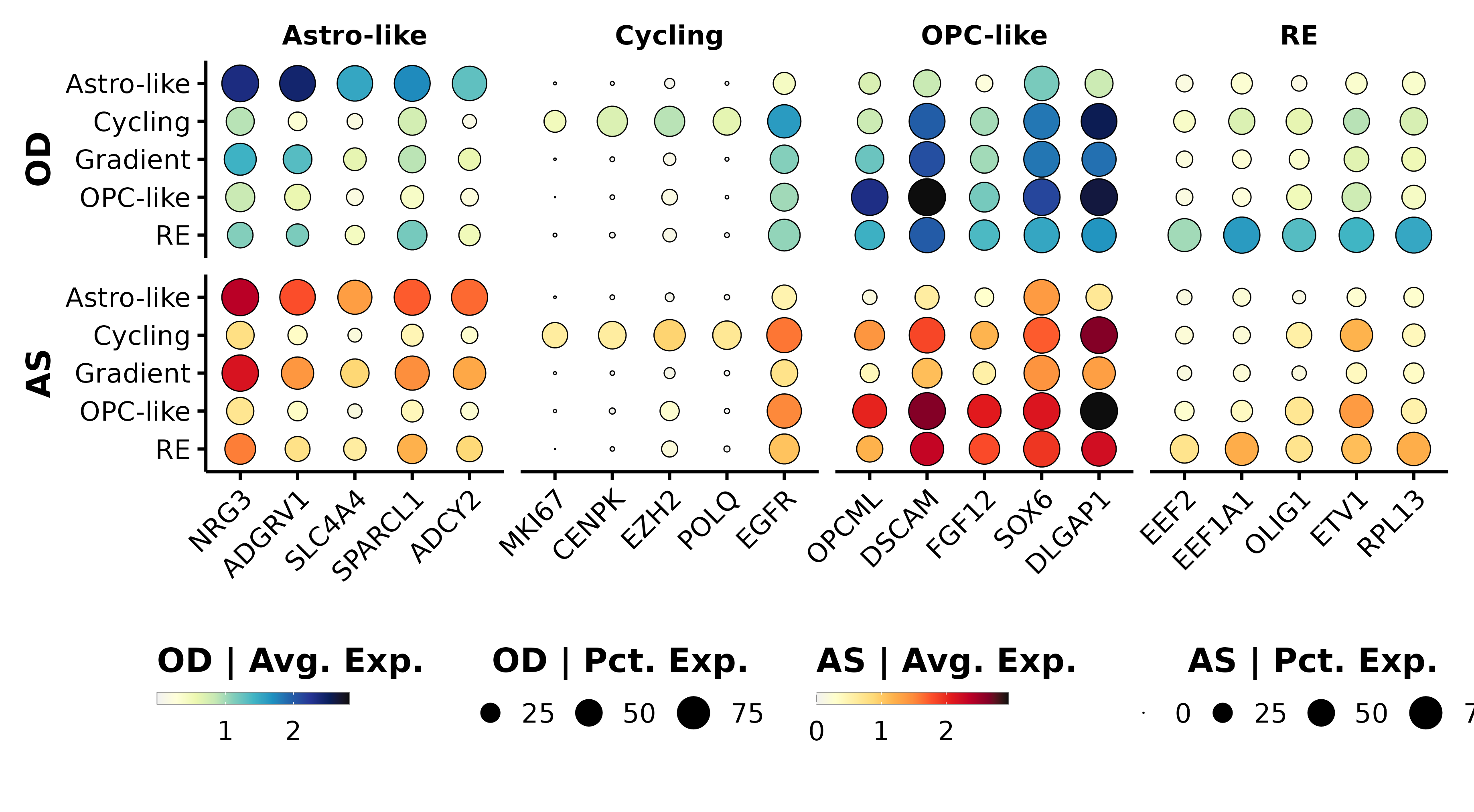
# Manually picked features from the NMF metaprograms.
features <- list("Astro-like" = c("NRG3", "ADGRV1", "SLC4A4", "SPARCL1", "ADCY2"),
"Cycling" = c("MKI67", "CENPK", "EZH2", "POLQ", "EGFR"),
"OPC-like" = c("OPCML", "DSCAM", "FGF12", "SOX6", "DLGAP1"),
"RE" = c("EEF2", "EEF1A1", "OLIG1", "ETV1", "RPL13"))
sample.oligo$relabelling <- factor(sample.oligo$relabelling,
levels = rev(c("Astro-like",
"Cycling",
"Gradient",
"OPC-like",
"RE")))
sample.astro$relabelling <- factor(sample.astro$relabelling,
levels = rev(c("Astro-like",
"Cycling",
"Gradient",
"OPC-like",
"RE")))
p1 <- SCpubr::do_DotPlot(sample.oligo,
features = features,
group.by = "relabelling",
assay = "SCT",
legend.title = "OD | Avg. Exp.",
legend.length = 8,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2,
dot.scale = 10,
font.size = 20,
axis.text.face = "plain") +
ggplot2::ylab("OD") +
ggplot2::theme(axis.text.x.bottom = ggplot2::element_blank(),
axis.ticks.x.bottom = ggplot2::element_blank(),
axis.line.x.bottom = ggplot2::element_blank(),
strip.background = ggplot2::element_blank(),
plot.margin = ggplot2::margin(t = 0, r = 10, b = 5, l = 10))
p2 <- SCpubr::do_DotPlot(sample.astro,
features = features,
group.by = "relabelling",
assay = "SCT",
sequential.palette = "YlOrRd",
legend.title = "AS | Avg. Exp.",
legend.length = 8,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2,
dot.scale = 10,
font.size = 20,
axis.text.face = "plain") +
ggplot2::ylab("AS") +
ggplot2::theme(strip.background = ggplot2::element_blank(),
strip.text.x.top = ggplot2::element_blank(),
plot.margin = ggplot2::margin(t = 0, r = 10, b = 0, l = 10))
p1$guides$size$title <- "OD | Pct. Exp."
p2$guides$size$title <- "AS | Pct. Exp."
p <- patchwork::wrap_plots(A = p1,
B = p2,
C = patchwork::guide_area(),
design = "A
B
C",
guides = "collect") +
patchwork::plot_annotation(theme = ggplot2::theme(legend.position = "bottom"))