p1 <- SCpubr::do_DimPlot(sample = sample,
group.by = "seurat_clusters",
font.size = 20,
raster = TRUE,
raster.dpi = 2048,
pt.size = 4,
legend.icon.size = 8,
legend.ncol = 2,
label = TRUE,
repel = TRUE,
legend.position = "none")
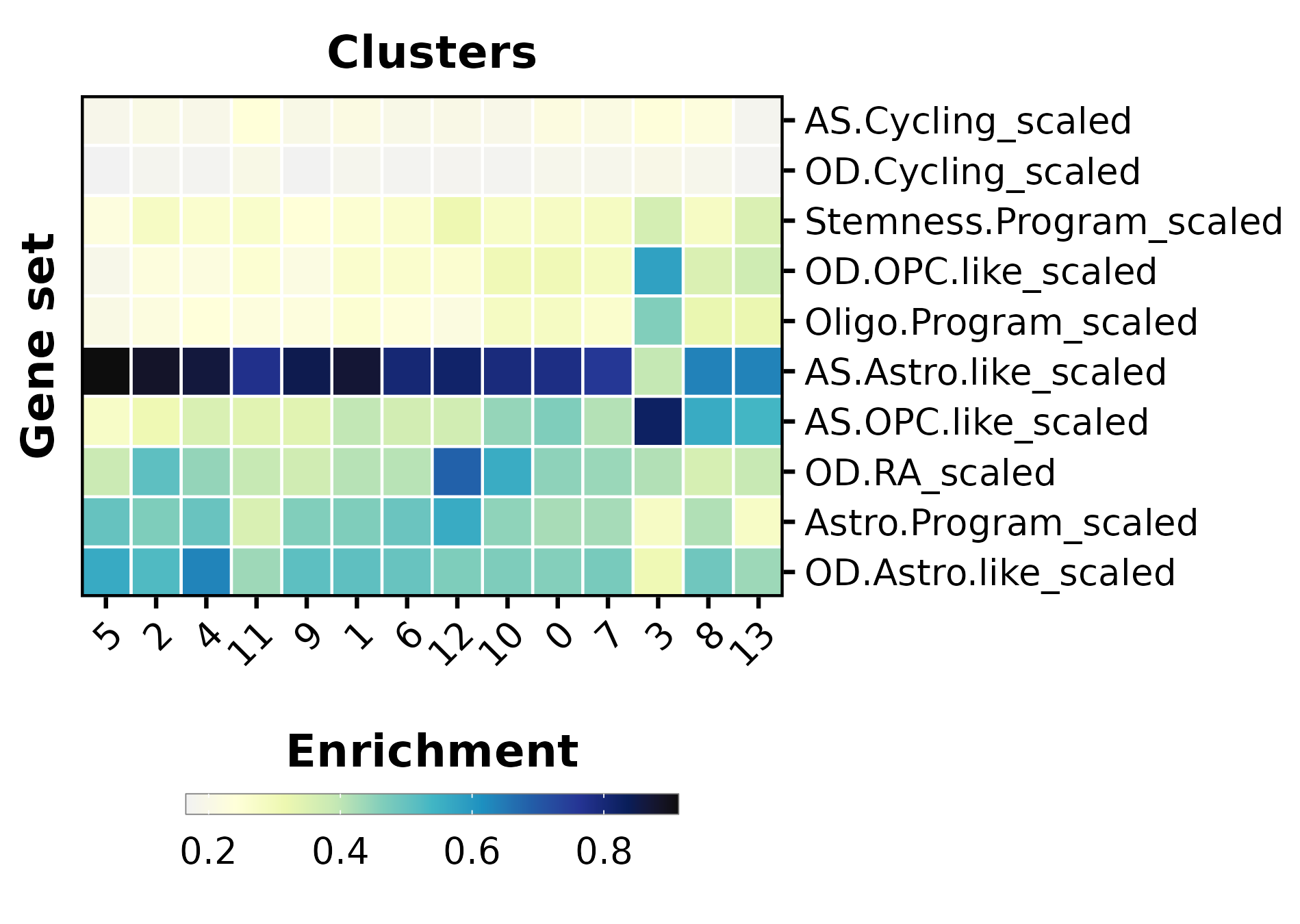
p2 <- SCpubr::do_EnrichmentHeatmap(sample = sample,
font.size = 16,
input_gene_list = markers,
group.by = "seurat_clusters",
flip = TRUE,
axis.text.face = "plain",
legend.length = 12,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2) +
ggplot2::labs(x = "Clusters", y = "Gene set")