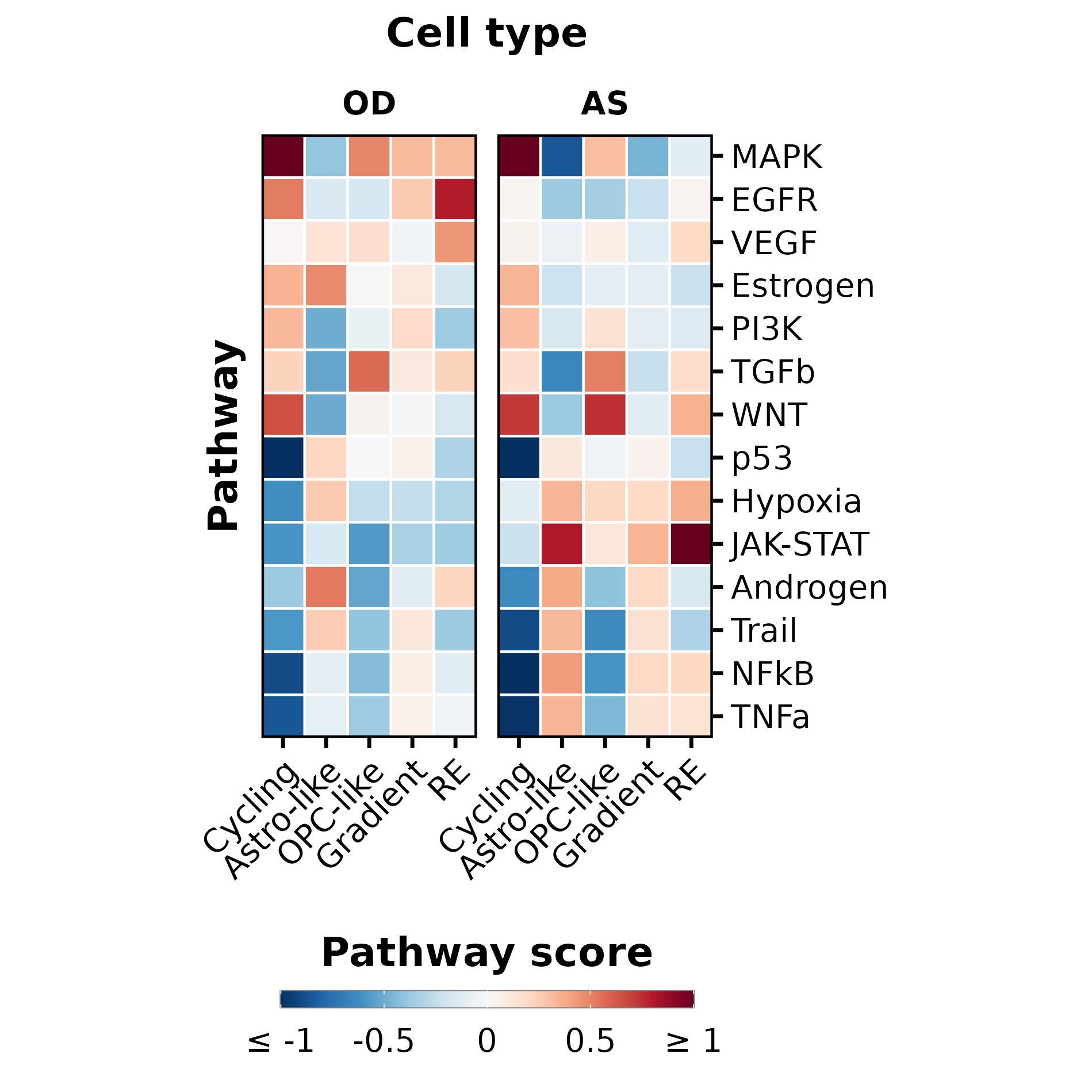
p <- SCpubr::do_PathwayActivityPlot(sample = sample,
activities = progeny_activities,
slot = "scale.data",
split.by = "subtype",
group.by = c("relabelling"),
min.cutoff = -1,
max.cutoff = 1,
flip = TRUE,
font.size = 17,
axis.text.face = "plain",
legend.length = 12,
legend.width = 0.5,
legend.framecolor = "grey50",
legend.tickcolor = "white",
legend.framewidth = 0.2,
legend.tickwidth = 0.2) +
ggplot2::ylab("Pathway") +
ggplot2::xlab("Cell type")
p$guides$fill$title <- "Pathway score"
p <- p + ggplot2::theme(plot.margin = ggplot2::margin(t = 0, r = 0, b = 0, l = 0))