out <- SCpubr:::compute_umap_layer(sample,
labels = colnames(sample@reductions[["umap"]][[]])[1:2],
border.color = "black",
pt.size = 1,
border.size = 2)
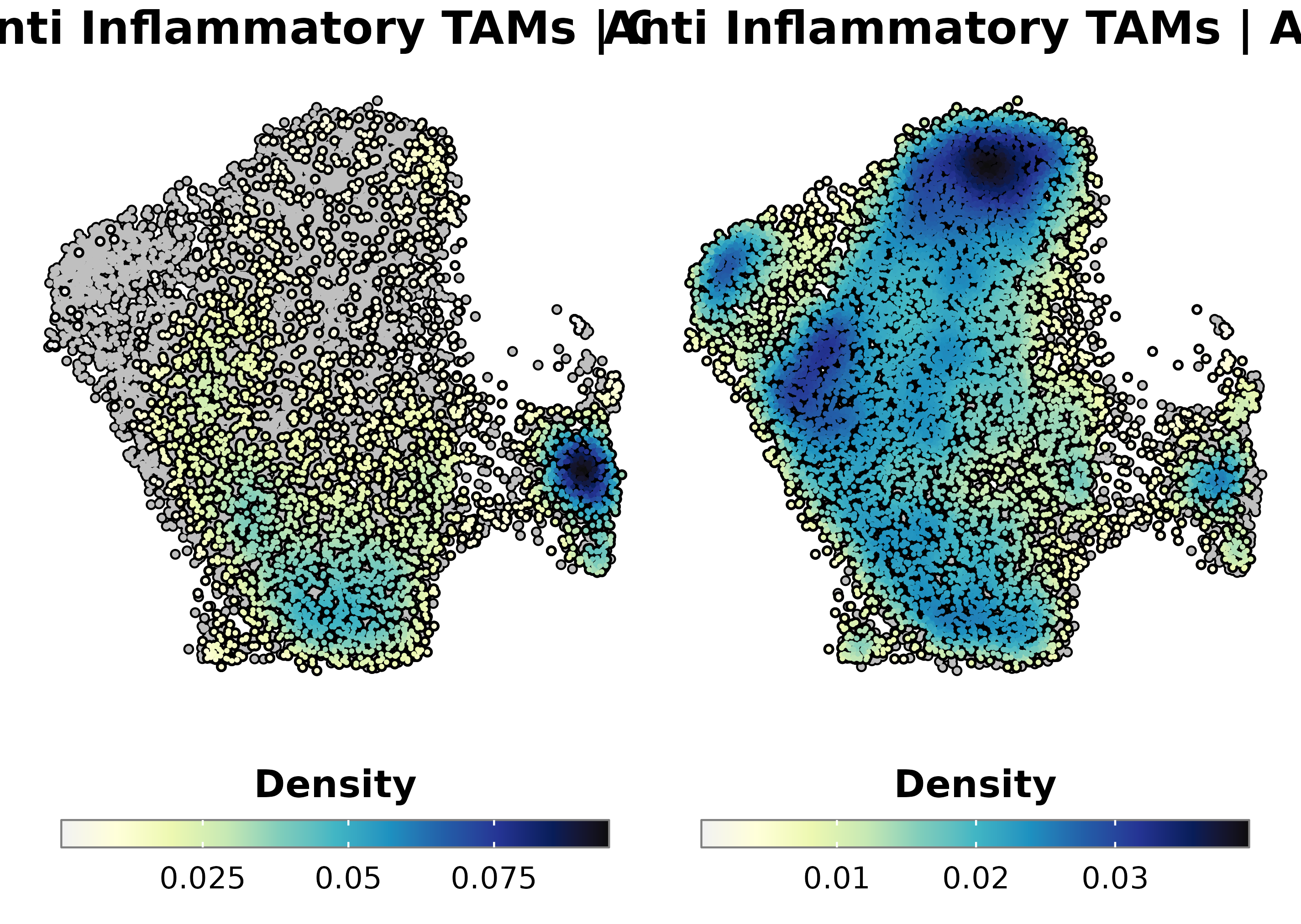
p1 <- SCpubr::do_NebulosaPlot(sample[, sample$subtype == "Astrocytoma"],
features = "Anti_Inflammatory",
font.size = 20,
use_viridis = FALSE,
plot.title = "Anti Inflammatory TAMs | AS",
sequential.direction = 1) + ggplot2::theme(plot.title = ggplot2::element_text(face = "bold", hjust = 0.5))
p1$layers <- append(out$na_layer, p1$layers)
p1$layers <- append(out$base_layer, p1$layers)
p2 <- SCpubr::do_NebulosaPlot(sample[, sample$subtype == "Oligodendroglioma"],
features = "Anti_Inflammatory",
font.size = 20,
use_viridis = FALSE,
plot.title = "Anti Inflammatory TAMs | OD",
sequential.direction = 1) + ggplot2::theme(plot.title = ggplot2::element_text(face = "bold", hjust = 0.5))
p2$layers <- append(out$na_layer, p2$layers)
p2$layers <- append(out$base_layer, p2$layers)
p <- p2 | p1